Promoting Interoperability and Standardization: Seven Bridges and GA4GH
The Global Alliance for Genomics and Health (GA4GH) is an international body set to create policies and promote technical standards to maximize interoperability among various stakeholders involved with genomics and healthcare-related data. Through its engagement in GA4GH, Seven Bridges actively works with platform development partners and industry leaders to develop standards that will facilitate interoperability. But how does this benefit our users?
Standardization can address the many issues of working across data ecosystems
Adopting standards is a crucial part of interoperability: the ability for different data ecosystems to share and make use of the information they contain. Before standardization, the process of retrieving data from and between ecosystems was not straightforward: oftentimes access tools which work in one environment do not apply to another, creating barriers between the user and the data needed for their analysis. As repositories of data are continuously filling with information of growing volume and complexity of data, an issue which will only grow with time, it becomes progressively more difficult for researchers to ensure that their data of interest is hosted in a manner that aligns with the cloud environment they seek to use for their data analysis. These issues are indicative of a lack of interoperability which could be remedied by the adoption of a unified set of standards.
If each different data ecosystem could all adopt a single, unified standard, it would allow the research community to reuse and share the work already completed, enhance interoperability, ensure scalability, and overall would lead to faster integration times. Specifically, researchers need to be able to import and export phenotypic and genotypic data, compute in a standard way across various environments, and also have a flexible means for authorization and authentication for the data across these environments. Ideally, this can all be done quickly and efficiently as possible, while ensuring safety and integrity. Towards this aim, GA4GH standards are immensely useful for democratizing access to data resources with a safe and secure approach.
Seven Bridges and GA4GH strive towards standardization and interoperability
Seven Bridges has historically made an effort towards the adoption of standards and efforts for interoperability: we first started monitoring and engaging with the GA4GH community in October 2018. Together, we created a strategy for the GA4GH Cloud Workstream. This Cloud Workstream helps researchers in the genomics and health science communities take full advantage of modern cloud environments. The initial focus of this effort is to “bring the algorithms to the data” by creating standards for defining, sharing, and executing portable workflows. Seven Bridges supports these standards for our platforms, and are building support for upcoming updates to these standards as well. We believe standards are important to be widely adopted across actors, institutions, and nations, in order to realize the goal of global outreach for research.
By being stakeholders in the GA4GH community, and through early and continued adoption of GA4GH policies, Seven Bridges acts to influence and drive standardization in directions most beneficial for our users. At Seven Bridges, we strive to be at the forefront of standardization in healthcare- and research-driven genomics. This applies to both the current state of the field, and our efforts to ensure continued growth and development on the Seven Bridges platforms in the future. In essence, Seven Bridges supports GA4GH engagement and efforts towards interoperability to better meet the needs of our users: allowing them to reach highly-distributed data in a standardized and efficient manner.
Facilitating standards adoption and pursuing a path towards interoperability will allow researchers to (1) access datasets that are hosted on other NIH cloud platforms from the primary platform that the researcher is using, and (2) co-analyze datasets that are hosted on different NIH cloud platforms. To facilitate this work, Seven Bridges has implemented and will continue to implement API and specifications from GA4GH.
Seven Bridges implementation of WES and DRS APIs
Adopting GA4GH standards would allow researchers to search for, find, and analyze data more easily across these diverse data ecosystems, such as between Gabriella Miller’s Kids First Pediatric Research Program (KF), CAVATICA, and the Cancer Genomics Cloud (CGC).
Currently, Seven Bridges provides support for the following standards on the CGC, CAVATICA, and NHLBI BioData Catalyst: the WES API and the DRS API.
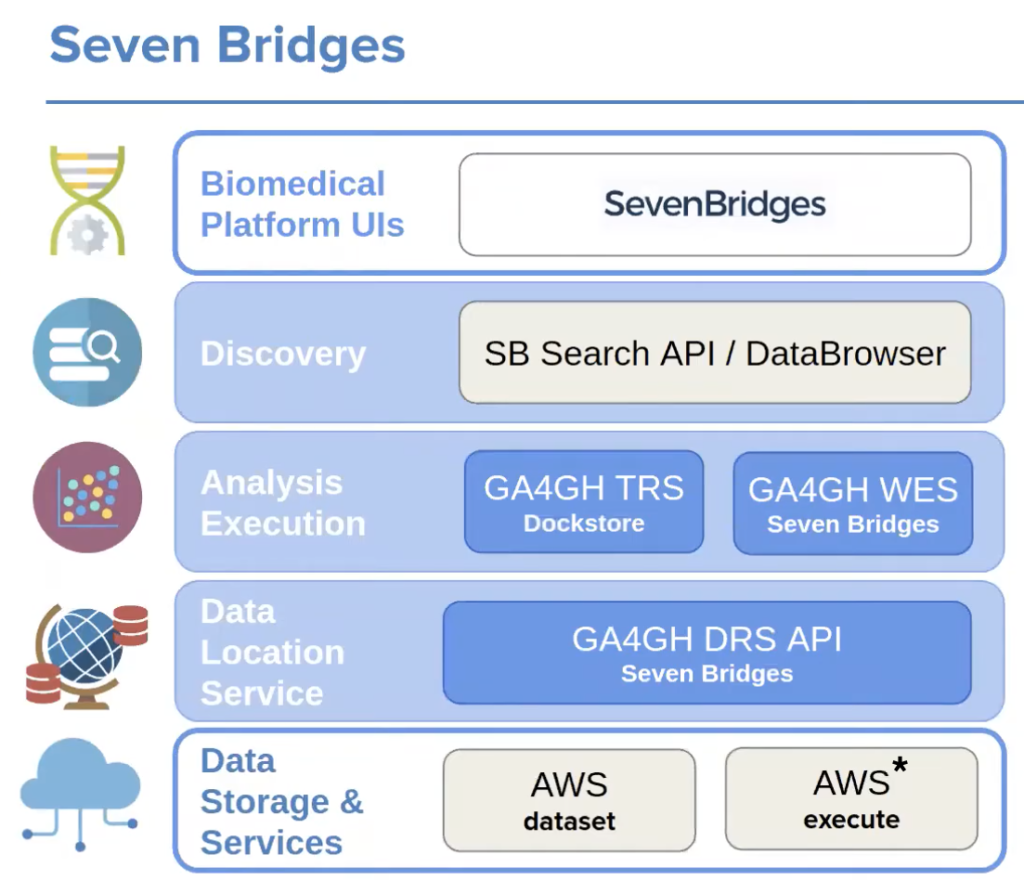
Figure 1: Overview of the Seven Bridges and GA4GH integration. The asterisk denotes that Seven Bridges also is compatible with Google Cloud Provider (GCP) for Data Storage and Services.
WES API
The Workflow Execution Service (WES) API describes a standard programmatic way to run and manage workflows. Having this standard API supported by multiple execution engines will allow users to run the same workflow using various execution platforms running on various clouds/environments. This WES API is a method by which users can submit workflow requests, run workflows, and monitor their performance from a dedicated API. Moreover, WES allows users to run workflows formatted in CWL on WDL on various types of platforms and cloud environments, freeing users from platform-specific or environment-specific constraints on the tools available to their research.
The following API paths are available as part of Seven Bridges implementation of WES API:
- Run a workflow
- Cancel a running workflow
- Get the status of the workflow run
- Get details of a workflow run
- List workflow runs
- Get service information
DRS API
The Data Repository Service (DRS) API provides a streamlined, user-friendly interface to data repositories so data consumers, including workflow systems, can access data in a single, standardized way, regardless of where the data is stored and how it is managed. Originally created by the GA4GH Steering Committee, the DRS API is a set of methods of accessing data that are not dependent on a specific type of cloud infrastructure. Think of the DRS API as a sort of data access “skeleton key,” removing obstacles between data access and data analysis, and ultimately promoting data sharing and proliferation. Overall, the DRS API provides a mechanism to read (and in the future, write) data objects across object stores in a cloud-agnostic way. Further integrating DRS with WES will facilitate the ability to search for a dataset of interest, then use that dataset as an input for subsequent workflows for analysis. Another advantage of utilizing DRS is that users can write one DRS client, and then have interoperability with any other DRS server in the world. This is especially valuable for integrating with new nodes as they come online over time. Of note, the National Cancer Institute (NCI) has highlighted Seven Bridges’ implementation of the DRS implementation at the Cloud Workstream, underscoring its value for the research community.
The following API paths are available as part of Seven Bridges implementation of DRS API:
- Get info about a DRS object
- Get a URL for downloading a file
Interoperability in Production
The 8th GA4GH Plenary occurred this past September, where Seven Bridges was featured in the Connection Demos together with three other stacks: DNAStack, Terra and Elixir on the Horizontal Demo (shown below). The work Seven Bridges has put forth in the Connection Demos has been referenced as one of the key takeaways from the conference, which is a testament to not only the efforts Seven Bridges has taken towards achieving the goal of interoperability and standardization, but also to the continued dedication to being pioneers in these efforts.
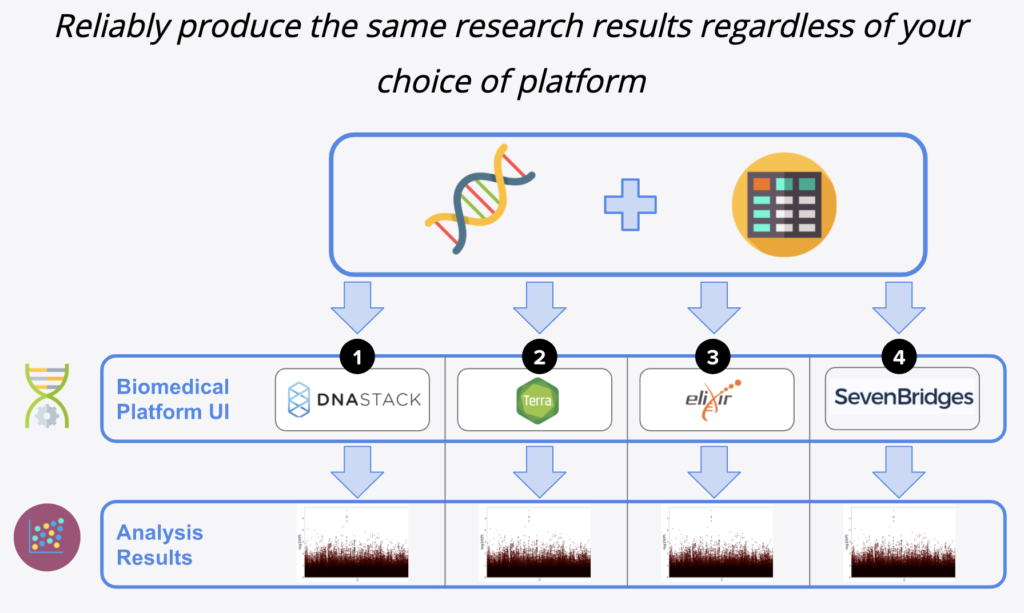
Figure 2: Interoperability in Production, from GA4GH’s Connection Demo series
Documentation for further reading
To learn more about Seven Bridges implementation of these standards on each of our platforms, please see the following links to the documentation below.