Develop, test and scale reproducible bioinformatics workflows with Rabix
Rabix: the toolkit for reproducible bioinformatics
This summer, our development team announced the public launch of Rabix: the open source toolkit for creating and running reproducible computational workflows. Rabix was founded to overcome the challenges inherent in running reproducible bioinformatics analyses at scale. Here we show how the combination of software containers, workflow language and Rabix gives bioinformaticians the tools to ensure recomputability, and make their analyses fully portable to support collaborative, reproducible bioinformatics at scale.
Software containers + Common Workflow Language = recomputability
Rabix utilizes software containers — a technology that makes it easy to share software across different compute environments. Software containers allow developers to package software into a discrete environment with its own virtual file system and dependencies. This reduces set up time in different computation environments, and minimizes errors when sharing and testing code. Technologies such as Docker have automated the process of creating and sharing software containers, spurring much broader adoption and lowering the technical barrier to entry.
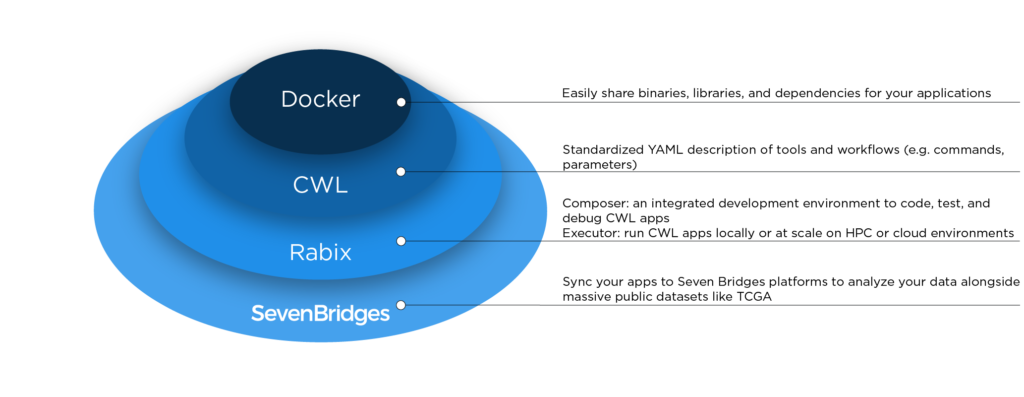
While software containers can contain all the ‘pieces’ of a bioinformatics analysis, they lack the instructions on how to use them to ensure full reproducibility of complex workflows. The Common Workflow Language (CWL) is a set of instructions that lets researchers and software developers describe how containerized bioinformatics applications are executed by standardizing description of tool inputs, outputs, parameters, and connections. CWL allows researchers to easily describe, share and publish complex bioinformatics workflows (for example, the CloudNeo workflow for identifying patient-specific tumor neoantigens), speeding the path to independent verification and adoption of these methods.
Workflow descriptions are a key component of computational reproducibility or recomputability. Computational reproducibility enables biomedical research organizations to record exactly which software or data generated a particular insight, such as a decision to target a specific gene or rule out an avenue of exploration. Computational reproducibility is an increasingly essential component of biomedical research, particularly if regulatory submissions involve complex bioinformatics workflows.
Rabix allows users to implement CWL to its full potential
While CWL is the emerging standard for reproducible bioinformatics, it has some limitations. First, manually coding workflow descriptions can be time consuming because complex bioinformatics workflows consist of many of tools and parameters, requiring hundreds of lines of code to describe them. Second, a CWL description of a workflow requires executors or workflow engines to interpret the specification and convert the description into computational jobs. Third these workflow engines require job scheduling optimizations to allow efficient computing on large volumes of data.
In response, we created Rabix—the open-source development project for creating and running CWL workflows. Rabix enables researchers and bioinformaticians to develop, test and debug CWL-described tools and workflows on their laptop, before sharing and executing them at large scale. It comprises two open source tools:
1. Rabix Composer: build reproducible bioinformatics workflows
The Rabix Composer (currently in beta release) is an integrated development environment for CWL. It provides rich visual and text editors to enable developers to easily create CWL descriptions. Rabix Composer provides a user interface for describing individual command line tools in CWL as well as a workflow canvas with interactive visualization that enable rapid assembly of tools into a workflow.
Rabix Composer’s visual tool editor provides form fields to comprehensively describe a tool’s commands and parameters. As you fill out the form fields, Composer dynamically generates and validates CWL code in the text editor. When you finish describing your tool, you can use the Rabix Executor (described below) for local execution of the CWL description. The Executor provides error logging to help you debug errors and iteratively develop your tools. By repeating this process for multiple tools and linking them into a workflow, local testing enables you to debug your application before you scale your analysis on an HPC or cloud environment.
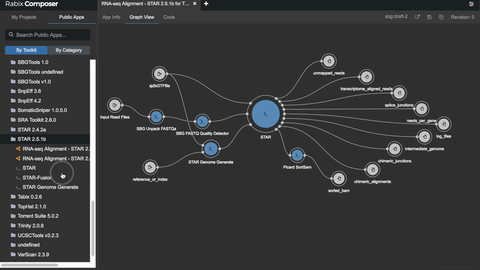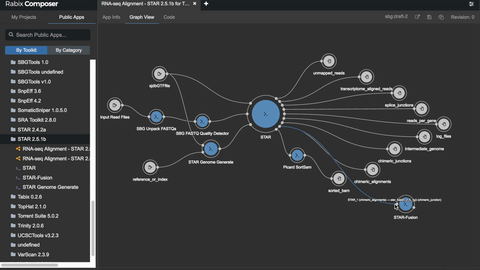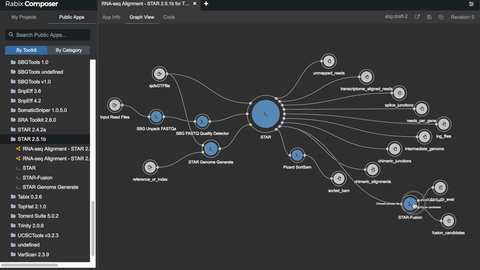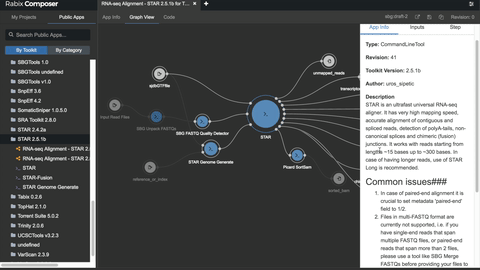
2. Rabix Executor: run CWL apps locally or at scale on HPC or cloud
The Rabix Executor (v1.0) provides a command line interface (CLI) for executing containerized software based on a CWL description in any Unix-based computational environment. The executor parses CWL descriptions into computational jobs, provides error logging, file organization, and job scheduling optimizations to allow efficient computing on large volumes of data. As the Executor runs in any Unix environment, you can easily deploy and share your locally developed workflows; successful computation on your laptop will translate to other computation environments.
The Rabix Executor has a modular design, so that developers can modify the major components as needed; for example, developers can modify the backend module to implement a queuing protocol of choice, to send analysis jobs to HPC clusters or cloud infrastructure. We have already leveraged this modularity to integrate Rabix Executor with the Global Alliance for Genomic Health (GA4GH) Task Execution Schema (TES) API for data analysis workflows, which aims to provide a standard open-source API for submitting analysis jobs to different backends. By integrating TES with the Rabix Executor, you can configure your local Rabix CLI to send analysis jobs to a remote TES API server instead of your laptop’s computation. The TES API can be easily plugged into most existing computation environments, including clusters and clouds.

Scaling analysis with Rabix and Seven Bridges
The Rabix Composer allows researchers to easily connect to any of the Seven Bridges research environments—including our commercial NGS analysis Platform, Cancer Genomics Cloud and Cavatica—to edit existing CWL applications locally, or to push new CWL applications to analyze data at scale, including massive public datasets like The Cancer Genome Atlas. The Rabix Executor is the workflow engine for all Seven Bridges environments, making locally developed applications fully portable to the cloud.
Rabix is free to use and open-source under the Apache 2.0 license. Your team can get started right now by downloading Rabix via rabix.io, including example tools and workflows demonstrating how to use CWL for reproducible and portable analysis.
Seven Bridges is a leader in the development of standards for reproducible bioinformatics analysis. Contact us to discuss how computational reproducibility can help accelerate your path from discovery to approval.
Many thanks from the Rabix Team to Patrick Grady for preparing the initial draft of this post.