What Makes TOPMed Datasets So Special?
Studies from the Trans-Omics for Precision Medicine (TOPMed) program are available for analysis on NHLBI BioData Catalyst. The TOPMed program, funded by the National Heart, Lung, and Blood Institute (NHLBI), part of the National Institutes of Health (NIH), focuses on data specifically for advancing science in the fields of heart, lung, blood, and sleep disorders. This program is generating a rich, multi-omic dataset with data from various -omics fields, such as genomics, proteomics, metabolomics, and transcriptomics, and is adding that information to pre-existing studies that have characterized thousands of phenotypic variables, including biochemical, physiological, clinical, behavioral, and anatomical measures. These TOPMed studies feature a great degree of racial and ancestral diversity compared to other genetic studies, which have typically focused on participants with European ancestry. Working with participants of other ancestries is important to advance scientific discovery for the many groups that have received significantly less (or no) attention from genetic research.
TOPMed currently includes whole-genome sequencing (WGS) of over 155,000 individuals, making it one of the largest, most diverse WGS datasets available to researchers. Furthermore, some TOPMed studies offer rich imaging datasets like that of COPDGene with 22 million CT scan images for about 10,000 study participants. The TOPMed program works alongside and complements various other programs, such as the All Of Us Research Program, The Million Veterans Program, and the NIH Database of Genotypes and Phenotypes (dbGaP). Overall, the TOPMed program is a powerful step toward developing precision medicine: using a patient’s individual genetic background and consideration of their environmental factors in order to formulate therapies and treatments specifically tailored for that individual.
TOPMed studies on NHLBI BioData Catalyst
NHLBI BioData Catalyst powered by Seven Bridges strives to facilitate access to TOPMed studies. The following TOPMed data is hosted on NHLBI BioData Catalyst today:
- Multi-sample VCFs for participants within 42 TOPMed studies. This includes multi-sample VCFs from Freeze8 as well as multi-sample VCFs from Freeze 5b. See the table below for the full list of hosted studies.
- CRAM files and single sample VCFs for all sequenced participants, the former of which are not available on dbGaP.
- Raw phenotype files for participants in TOPMed studies, providing clinical information such as BMI and lipids levels. In some cases, these data are in different dbGaP accessions.
- Single sample VCFs for subjects included in Freeze5.
Seven Bridges is currently working with the NHLBI BioData Catalyst consortium to onboard additional studies and data that have been released on dbGaP as part of TOPMed Freeze 8.
Working with the TOPMed studies on the cloud
You can view the available TOPMed studies in the platform Data Browser feature and select studies to query. You can search for specific file types in the Data Browser as well as by study consent group. The following data types are available for the hosted TOPMed studies:
| Data Type property in Data Browser | Genomic data file type |
| Aligned Reads | CRAM files |
| Simple Germline Variation | Single sample VCF files |
| Unharmonized Clinical Data | Raw phenotype files from dbGaP |
| Variant Call | Multi-sample VCF files |
You can find these files by searching the “File” entity and using the “Data Type” property in the Data Browser, as shown in the image below:
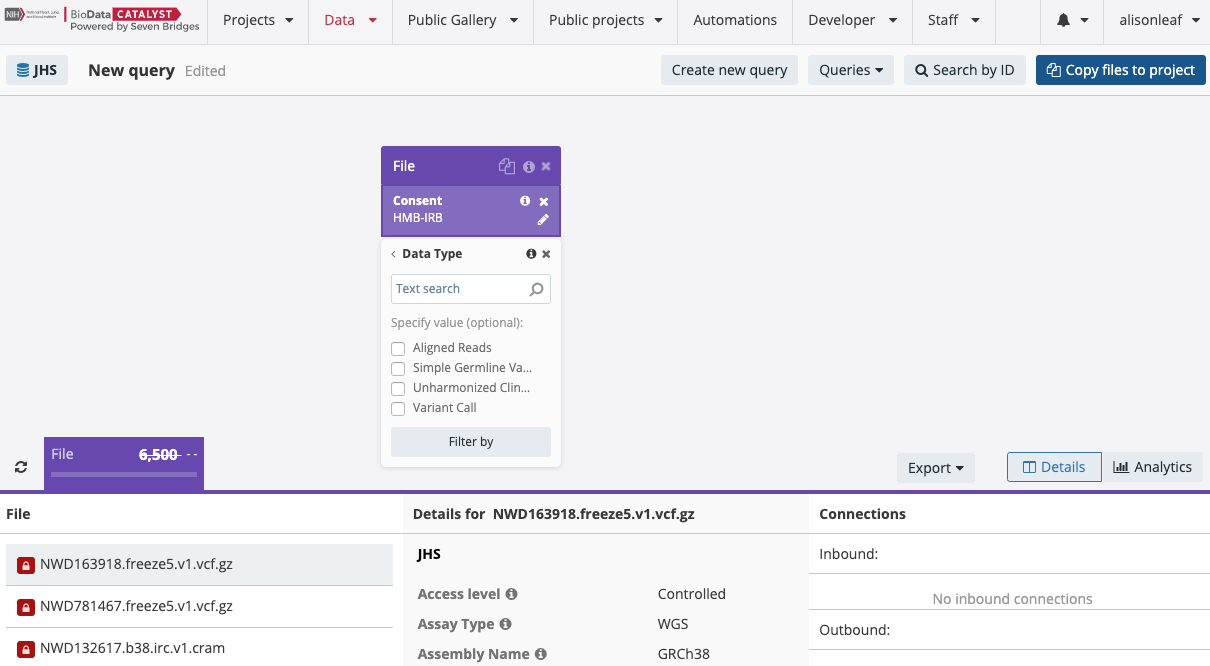
You can search specifically for the Freeze8 multi-sample VCFs by checking the box next to “Variant Call” in the Data Type property and then subsequently adding the “Freeze” property to the search as shown below:
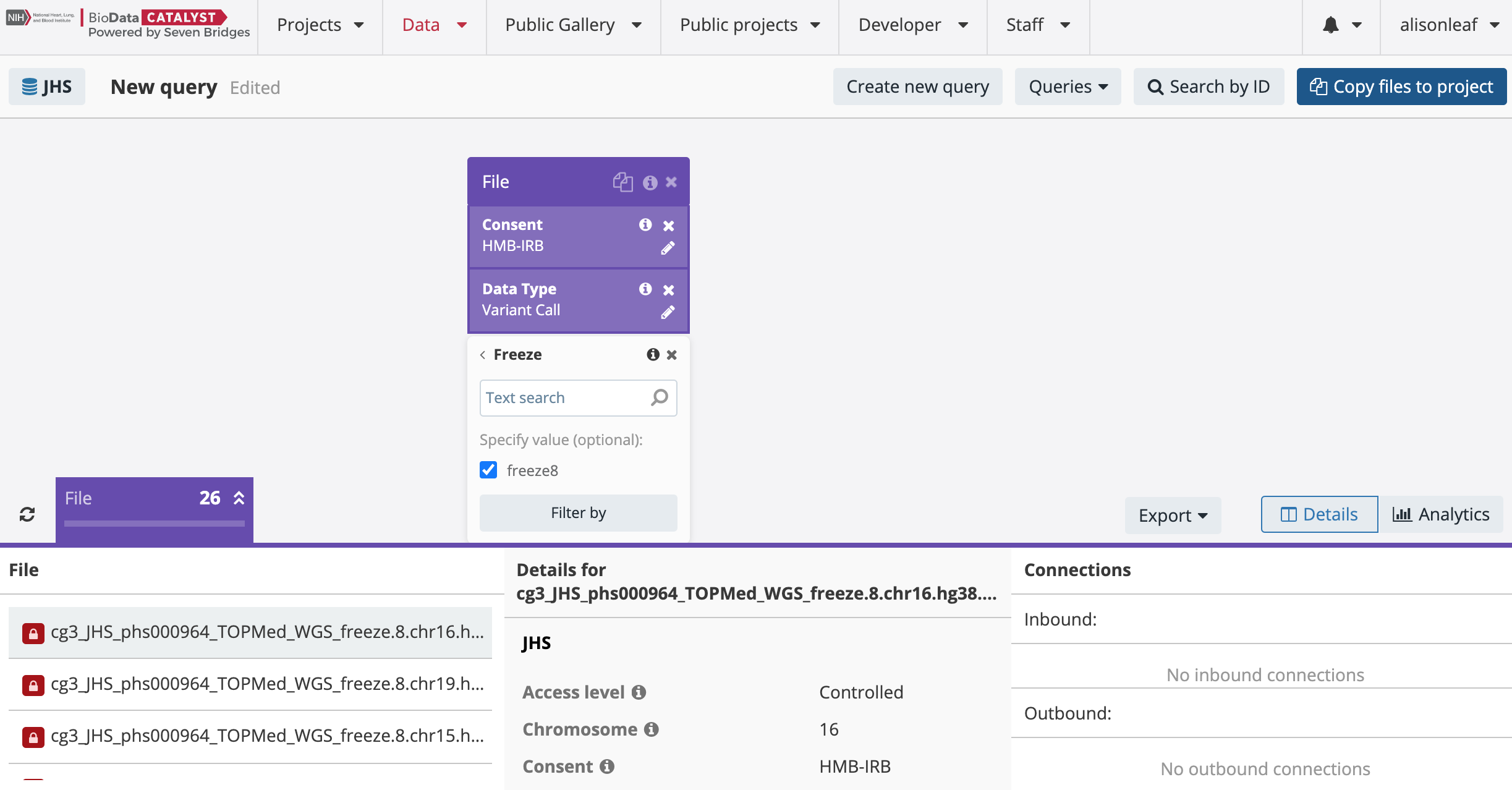
A search for Freeze8 multi-sample VCFs from a particular study and consent group will result in VCF files for each chromosome. This is in contrast to dbGaP, where the released Freeze8 data is available in tar bundles. Please note that the Freeze5 multi-sample VCFs are in tar bundles for each consent group within a TOPMed study. These tar files can be decompressed on the platform using the Seven Bridges Decompressor App by searching “decompressor” in the Public Gallery of Apps.
The Data Browser exposes only open metadata from the TOPMed studies for search, so all researchers are able to do the same searches and see the existence of all files. However, only users with appropriate dbGaP approval can add files to their project to use in an analysis. A service within NHLBI BioData Catalyst programmatically reads user permissions from dbGaP to determine if a user can access particular files on the system. This service recognizes if a user has a Data Access Request in dbGaP for TOPMed data or if a user is set as a “dbGaP downloader” for a particular dataset. Therefore, these are the two mechanisms for getting access to TOPMed studies on NHLBI BioData Catalyst. Please note that phenotype and genotype data for some studies are in different dbGaP accessions. More information is available on the Data page of the NHLBI BioData Catalyst website.
To learn more about how to get started working with the TOPMed studies on NHLBI BioData Catalyst powered by Seven Bridges, take a look at the Getting Started Guide which describes how to create an account, set up projects, run analyses, and find the TOPMed data in the Data Browser.
Further information on the hosted datasets can also be found on the Seven Bridges Documentation section “Datasets Hub”.
For more information on which TOPMed studies and parent studies are offered, including their phs identification numbers used by dbGaP, please see the list here.
Be sure to receive late-breaking updates from Seven Bridges and follow us on LinkedIn and Twitter.