Seven Bridges is the biomedical data analysis company specializing in making data useful. We empower researchers to work with the world’s largest and richest genomic data sets, alongside other biomedical and clinical data. Seven Bridges combines expertise in data science, bioinformatics and population-scale genomics to accelerate research for cancer, drug development and precision medicine.
Making data usable, not just available
As the amount of biomedical data increases worldwide, only a small fraction of it is immediately usable. Managing a large data set for research is time-consuming, costly, and requires skilled personnel. Seven Bridges does this work, freeing your organization’s resources to focus on research.
Our bioinformaticians download and curate the world’s largest genomic data sets including The Cancer Genome Atlas (TCGA) and Cancer Cell Line Encyclopedia (CCLE). We associate every data resource with a sophisticated metadata record that enables fast queries (both visual and programmatic), provenance tracking, and integration with future data sets. Based on semantic technologies, our metadata engine harmonizes ontologies to enable data to be explored and functionally organized in a way that makes sense to researchers.
The Platform for useful data
The Seven Bridges Platform enables secure bioinformatics research on public clouds and on-premise compute infrastructures. It acts as a central hub for teams to store, analyze and jointly interpret bioinformatics data. The Platform co-locates analytic tools and workflows with petabyte-scale genomics datasets and manages compute resources on demand.
The Platform centrally stores data for analysis across your organization, with metadata that enables it to be found, used and integrated with other data sources. Users of the Platform can immediately work with massive public datasets, such as TCGA and CCLE, alongside reference files and their own data. We also make available over 200 of the most popular tools and workflows for bioinformatics analysis, pre-loaded and kept up to date by our bioinformaticians.
Case study: Making The Cancer Genome Atlas usable
The Cancer Genome Atlas (TCGA) is the world’s largest and richest collection of genomic data, comprising 2.5 Petabytes of multi-omic data from over 11,000 patients, along with accompanying clinical and technical information. Downloading TCGA takes at least several weeks even with an optimized network connection. Once downloaded, integrated analysis of this data remains out of reach for most researchers, and is resource-intensive even for those with access to the largest institutional computation clusters.
In 2014, the US National Cancer Institute selected Seven Bridges to develop the Cancer Genomics Cloud, to solve the data accessibility and usability challenges that restricted the insights gained from TCGA. Recognizing that making data available is not the same as making it usable, our bioinformaticians worked over a period of 3–4 months to understand the available TCGA metadata and optimize its nomenclature. Alongside this, it took approximately 2 months to import the TCGA files to our Platform (>1 PB of data imported to an Amazon Web Services bucket). Finally, our team spent a further 3 months developing methods for accessing and exploring the data, including an intuitive visual data browser (see Figure).
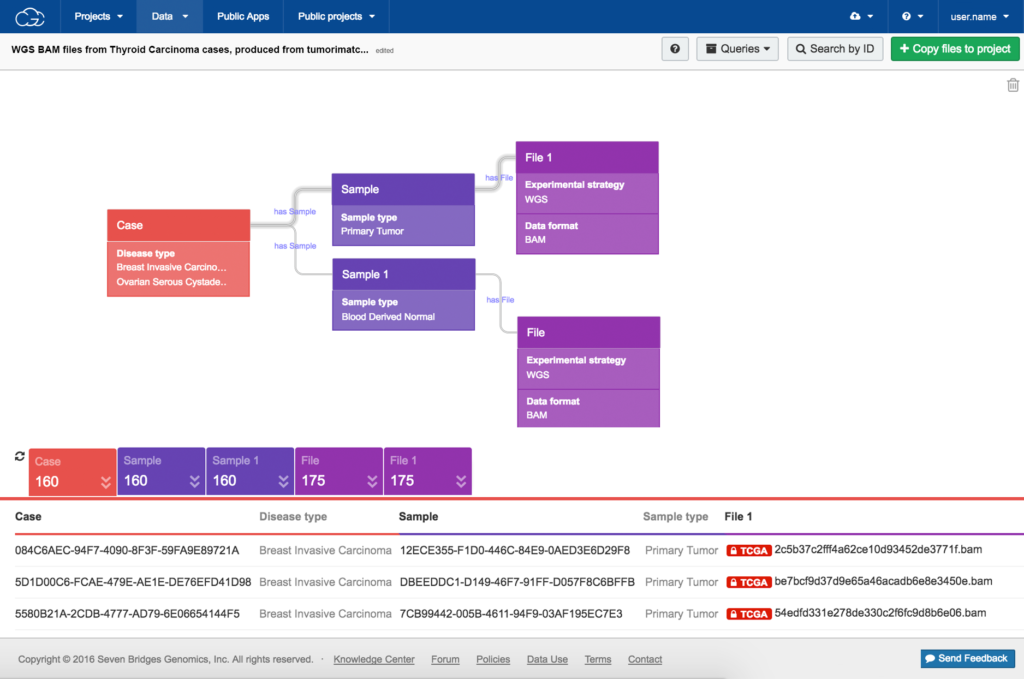
A visual interface lets researchers with diverse expertise build advanced queries to quickly find explore and use data of interest.
The Cancer Genomics Cloud launched in February 2016, and is currently used by over 1,200 researchers around the world. By using Seven Bridges, these researchers now immediately work with TCGA, in conjunction with other biomedical data, to make discoveries.